My First Foray into Live Applications Genetic Algorithms

In this blog post I will give a summarized background for the works Spawn 1 for piccolo (here shown with visuals) and Anemorpha for flute.
On wanting to try this application of genetic algorithms, my instinct was to turn to Python, as I knew there were many ways to implement this program and send streams of data in real time into Max through OSC. Indeed, there are whole libraries dedicated to these algorithms in Python. I started with this tutorial and was quite intrigued. However, since I am under some time constraint, I decided to defer this method and see if Max's ability to do math would allow me to strip down the process and run everything directly in Max. This turned out to be the case and here are the results.
My implementation of fitness descent is of course massively simplistic. However, that gives it some interpretability: I have used the same patch to conceptualize the evolution of a population (Spawn for piccolo) as well as an imagined life cycle of a wind-borne creature that undergoes metamorphoses (Anemorpha for flute).
To experiment quickly I constrained myself to a space that is low dimensional, smooth, and works toward a specific optimization (grow larger). For this, I set two "populations" of genes, a current one and a candidate one, to "compete" through a 2D inverted Gaussian function. This is not a "true" genetic algorithm, but I am not attempting to model evolutionary dynamics as a whole. Rather I am using it as a fitness function to test "adapted" behavior. Reflecting the two dimensional world, each "gene" is represented as a 2D vector.
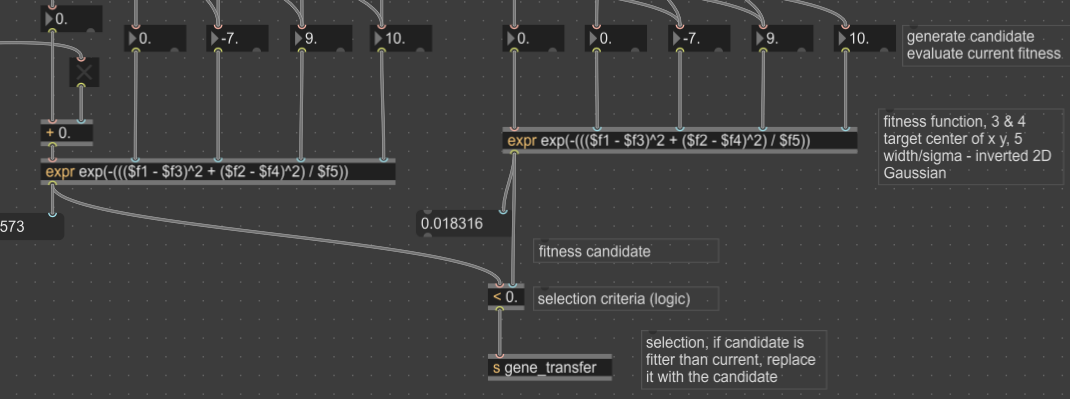
The live aspect: using pitch confidence (through fluid.pitch), I can influence the mutation strength of the candidate's "gene", and therefore the likelihood that it will transfer its "gene". The current "gene" (2D vector) is scaled values between 0 - 127, mapped onto an xy pictslider, and regressed (fluid.mlpregressor) to the parameters of a ring modulator. Additional processing, delay transpose and concatenative synthesis, is triggered by the motion of the xy slider. Future versions will include these processes in the regression, if only for the sake of comparison between regressed parameters and timed, linear behavior.
For experimental purposes of rendering, mixing down, and uploading, I kept the duration of these works short; in the set-up shown, the time it takes to reach its maximum size (arbitrarily decided my me) is ca. 4 minutes. Scaling this into a longer work will involve tweaking the numbers, mutation rate, and scalings to the xy pictslider. It will also likely involve additional processing elements to make a more variegated soundscape.
Where this live implementation succeeded: a true, nonlinear, incremental trajectory from one corner to the diagonal opposite on the xy pictslider was achieved. (This was planned in order to reflect the goal of "growth" in the "genes".) Each time the trajectory was unique in that the "steps", controlled by "gene" transfer were incremental, but different always in their timing and size. How this would compare to a "drunk walk" implementation would be interesting.
Where this live implementation "failed": it was not always clear how much my pitch confidence actually changed the mutation strength. At the beginning of the piece, it seemed I had some influence, but towards the end of each cycle, the incrementation often seemed to slow regardless of what I did, likely due to the nature of the fitness evaluation. More experimentation is needed here.